Never Test for Normality
Way back in 2013, I wrote this blog post explaining why you should never use a statistical test to check whether a sample came from a Gaussian distribution. I argued that data from the real world never come from a Gaussian distribution, or any other simple mathematical model, so the answer to the question is always no. And there are only two possible outcomes from the test:
- If you have enough data, the test will reject the hypothesis that the data came from a Gaussian distribution, or
- If you don’t have enough data, the test will fail to reject the hypothesis.
Either way, the result doesn’t tell you anything useful.
In this article, I will explore a particular example and demonstrate this relationship between the sample size and the outcome of the test. And I will conclude, again, that
Choosing a distribution is not a statistical question; it is a modeling decision. No statistical test can tell you whether a particular distribution is a good model for your data.
For the technical details, you can read the extended version of this article or run this notebook on Colab.
I’ll start by generating a sample that is actually from a lognormal distribution, then use the sample mean and standard deviation to make a Gaussian model. Here’s what the empirical distribution of the sample looks like compared to the CDF of the Gaussian distribution.
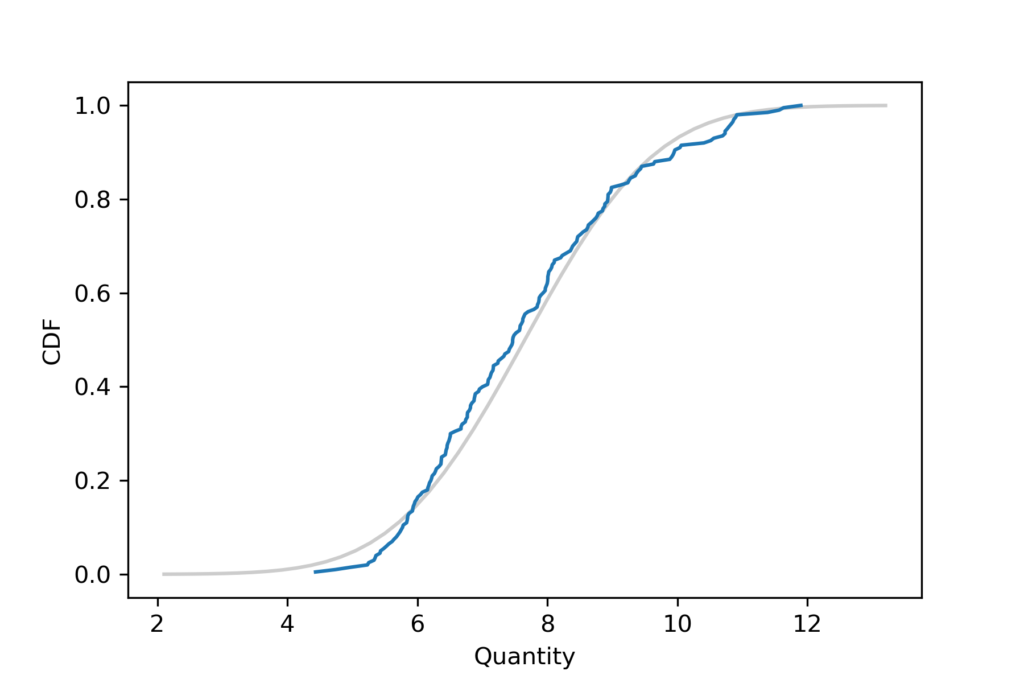
It looks like the Gaussian distribution is a pretty good model for the data, and probably good enough for most purposes.
According to the Anderson-Darling test, the test statistic is 1.7, which exceeds the critical value, 0.77, so at the 5% significance level, we can reject the hypothesis that this sample came from a Gaussian distribution. That’s the right answer, so it might seem like we’ve done something useful. But we haven’t.
Sample size
The result from the A-D test depends on the sample size. The following figure shows the probability of rejecting the null hypothesis as a function of sample size, using the lognormal distribution from the previous section.
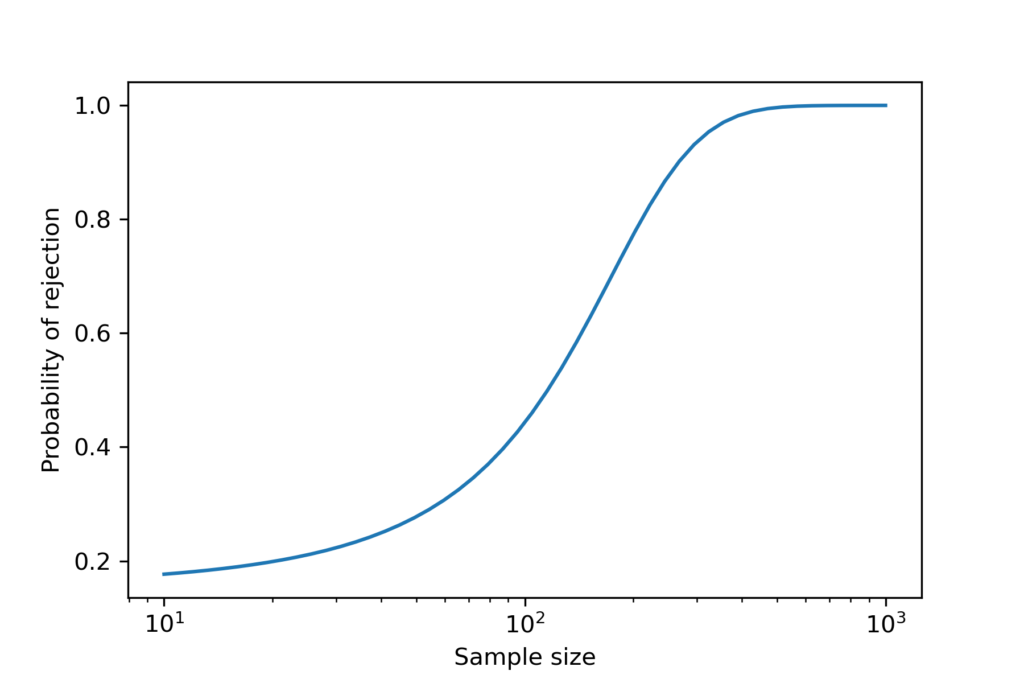
When the sample size is more than 200, the probability of rejection is high. When the sample size is less than 100, the probability of rejection is low. But notice that it doesn’t go all the way to zero, because there is always a 5% chance of a false positive.
The critical value is about 120; at that sample size, the probability of rejecting the null is close to 50%.
So, again, if you have enough data, you’ll reject the null; otherwise you probably won’t. Either way, you learn nothing about the question you really care about, which is whether the Gaussian model is a good enough model of the data for your purposes.
That’s a modeling decision, and no statistical test can help. In the original article, I suggested some methods that might.